This chapter was published as Uruñuela et al. (2020).
This chapter proposes the implementation of a subsampling approach based on stability selection that avoids the choice of any regularization parameter for hemodynamic deconvolution with sparsity-promoting regularized least squares estimators. The proposed method is implemented to operate with the Sparse Paradigm Free Mapping (SPFM) algorithm and its performance is evaluated on real fMRI data and compared with both the original SPFM algorithm, which used model selection criteria to select the parameters, and a conventional analysis with a general linear model (GLM) that is aware of the temporal model of the neuronal-related activity. The results demonstrate that SPFM with stability selection yields activation maps with higher resemblance to the maps obtained with GLM analyses and offers improved detection of neuronal-related events over SPFM, particularly in scenarios with low contrast-to-noise ratio.
Introduction¶
In the preceding chapter, deconvolution approaches were discussed in the context of functional magnetic resonance imaging (fMRI) data analysis as they offer the remarkable ability to estimate neuronal-related activity without the need for prior information on the timings of the blood oxygenation level-dependent (BOLD) events via a linear time-invariant model (i.e., a forward model of the BOLD response) that is then inverted by means of regularized least-squares estimators to deconvolve the neuronal-related activity at each voxel Gitelman et al., 2003Khalidov et al., 2011Karahanoğlu et al., 2013Hernandez-Garcia & Ulfarsson, 2011Gaudes et al., 2010Gaudes et al., 2013. In particular, the sparse Paradigm Free Mapping (SPFM) method Gaudes et al., 2013, which is the basis of this work, employs sparsity-promoting regularization terms based on the L1-norm of the estimates (e.g., using the LASSO or the Dantzig Selector). Importantly, inverse problem solving is linked to a dilemma that has yet to be solved: the selection of the regularization parameters that yield accurate estimates. As described in the previous chapter, methods based on model selection criteria after the computation of the entire regularization path Gaudes et al., 2013 or iterative procedures so that the variance of the residuals after deconvolution is equal to a prior estimate of the noise variance Karahanoğlu et al., 2013 have been previously used in the literature for parameter tuning due to their reduced computational cost. Yet, these methods offer no information about the appropriateness of the selected parameters.
This chapter proposes the use of the subsampling approach of stability selection Meinshausen & Bühlmann, 2010 with the SPFM algorithm Gaudes et al., 2013 to avoid the choice of any regularization parameter and account for the likelihood of the different possible estimates in the regularization path. Although stability selection has been previously proposed in fMRI data analysis, for example in the estimation of functional connectivity matrices from partial correlations with sparse estimators Ryali et al., 2012 and to detect change points in time-varying functional connectivity with the graphical lasso Cribben et al., 2013, its application for the deconvolution of the fMRI signal is innovative. Further, this chapter implements a novel procedure that enables to benefit from the computational speed of the least angle regression algorithm Efron et al., 2004 in combination with the robustness of stability selection.
This chapter uses a modification of the original SPFM formulation called block model --and introduced in Chapter 2: Synthesis-Based and Analysis-Based Hemodynamic Deconvolution for fMRI-- that computes estimates of the innovation signal of the neuronal-related signal (i.e., defining its changes) Karahanoğlu et al., 2013Cherkaoui et al., 2019Uruñuela et al., 2023, rather than the signal itself. The block model formulation fits the data used in this study better as it improves the estimation of neuronal-related events with long, sustained activity Karahanoğlu et al., 2013Cherkaoui et al., 2019Uruñuela et al., 2023 that cannot be adequately described by conventional spike-like models Khalidov et al., 2011Gaudes et al., 2010Gaudes et al., 2013. Nevertheless, the proposed stability selection procedure can be readily implemented for the spike model. The chapter is organized as follows: in Signal Model and Deconvolution with Stability-Based Paradigm Free Mapping the signal model and the stability-based SFPM algorithm are introduced; in Methods, the results of applying this new algorithm on experimental fMRI data are presented and compare them to the previous SPFM algorithm.
Signal Model and Deconvolution with Stability-Based Paradigm Free Mapping¶
For the sake of completeness, this section revises the signal model for hemodynamic deconvolution, which was already described in Chapter 2: Synthesis-Based and Analysis-Based Hemodynamic Deconvolution for fMRI. In fMRI data analysis, the signal of a voxel is commonly modelled as the convolution of an underlying neuronal-related signal with the hemodynamic response function (HRF) , plus a white noise component: , or in a discrete-time matrix notation. Typically, the neuronal-related signal is represented as a train of Dirac impulses at the fMRI timescale associated with the experimental design. This model of the neuronal-related signal has been adopted by previous deconvolution algorithms Hernandez-Garcia & Ulfarsson, 2011Gaudes et al., 2010Gaudes et al., 2013 relying on regularized least-squares estimators as follows:
where the -norm penalizes the amplitude of the coefficients of the neuronal-related signal, e.g., (i.e., ridge regression) and (i.e., LASSO) were employed in Gaudes et al., 2010 and Gaudes et al., 2013, respectively. Instead of the on/off pattern described by Dirac impulses, the neuronal-related signal can also be represented as a piecewise constant signal in terms of its innovation signal (i.e., its first derivative in time). Defining where corresponds to the discrete integration operator Cherkaoui et al., 2019, the signal model can be written as:
where , , and is the Toeplitz convolution matrix with shifted HRFs, where is the number of observations of the fMRI signal. The signal will represent those instances when significant changes in the neuronal-related activity occur. Since the innovation signal is sparser than the neuronal-related signal , it is also a more adequate representation if the temporal deconvolution of the fMRI time series of each voxel is performed with L1-norm regularized estimators as follows:
Combining Stability Selection with Least Angle Regression¶
An appropriate choice of the regularization parameter λ in Eq. 1 or Eq. 3 is crucial for appropriate hemodynamic deconvolution. Several techniques to select it have already been proposed, such as based on the Bayesian Information Criterion Gaudes et al., 2013. However, these techniques do not provide a solution that is robust regardless of the different characteristics the data may show (e.g., signal-to-noise ratio, occurrence and duration of neuronal events).
This problem can be overcome by implementing a novel procedure based on the stability selection approach Meinshausen & Bühlmann, 2010. This procedure generates surrogate datasets where the original voxel time series is randomly subsampled to retain 60% of its time points. Then, the optimization problem in Eq. 3 is solved for each surrogate dataset, where the model matrix is subsampled accordingly. Then, the stability paths of the signal for each surrogate and each time point (i.e., ) are computed, which represent the probability of the coefficient being non-zero for a given λ. Originally, the stability selection approach operates by solving Eq. 3 for a predefined set of λ values, for example by means of the fast iterative shrinkage thresholding algorithm (FISTA) Beck & Teboulle, 2009. Instead, this chapter proposes to use the least angle regression (LARS) algorithm Efron et al., 2004, which computes the entire regularization path for an optimal decreasing set of λ values and is faster than FISTA Beck & Teboulle, 2009 for our purposes. Then, for each surrogate, the estimate at the regularization parameter and time point is binarized as if or otherwise. To overcome the fact that solving Eq. 3 with the LARS algorithm will generate a different set of λ values in each subsampled surrogate, a new set of λ values is created. This new set contains all of the regularization parameters from all of the surrogate-specific regularization paths in decreasing order. The coefficients remain 0 or 1 according to the preceding value of corresponding to the surrogate-specific regularization path computed by LARS. This step allows us to calculate the probabilities that construct the stability paths as the ratio of surrogates where each coefficient is different from 0 at each λ.
Furthermore, unlike in the original stability selection procedure that sets a given probability threshold to select the final set of non-zero coefficients, this implementation calculates the area under the curve (AUC) of the stability paths of each coefficient as follows:
where
represents the selection probability of coefficient for a particular choice of the regularization parameter , and is the total number of regularization parameters from all of the LARS regularization paths. Hence, the voxelwise time series reveals the most prominent coefficients indicating the probability of activation at each time point.
Thresholding and Debiasing¶
Afterwards, the AUC time series for each voxel are thresholded to identify those instances with high probability of a significant change in neuronal-related activity occurring. This threshold is based on a given percentile (or maximum) of the AUC values in a region of interest where no BOLD signal changes related to neuronal activity are assumed to occur (or can be detected). For example, the threshold can be set to the 99th percentile of the AUC values of deep white matter (DWM) voxels (see Results and Discussion).
Finally, it is recommended to remove the bias in the estimates of the neuronal-related signal owing to the L1-norm regularization term. For the signal model in Eq. 1 used in the original SPFM approach Gaudes et al., 2013, a debiased estimate of can be obtained by solving a least squares problem with a selection of non-zero AUC coefficients.
Rather, in the signal model with the innovation signal, the selected non-zero coefficients of are used to define a matrix whose columns are activation segments with piecewise constant unit between two non-zero coefficients of Zoller et al., 2019. A final debiased estimate of is obtained by solving the following least squares problem:

Figure 1:Flowchart of the stability-based SPFM algorithm.
Figure 1 illustrates the flowchart of the proposed stability-based SPFM algorithm.
Methods¶
The operation of the proposed stability-selection SPFM algorithm is illustrated in a dataset collected on a healthy subject in a 7T MR scanner (Siemens) using a 32-channel receive transmit coil. The subject was scanned under a Cleveland Clinic Institutional Review Board approved protocol (QED, Cleveland, OH). A volumetric MP2RAGE image was acquired for anatomical visualization. Two fMRI datasets were acquired with a simultaneous multislice EPI sequence (MB factor = 3, TE = 21 ms, field of view = ) at TR = 2800 ms (, flip angle = 55º) and 500 ms (, flip angle = 70º). For both TRs, the subject performed finger tapping events with the right index and thumb fingers every 45 s, where a single tap was performed in the first 6 minutes, or 10 taps quickly for the remaining 4 minutes. The onsets and durations of the paradigm are shown as gray vertical lines in Figure 2 (a) and Figure 3 (a).
Data preprocessing comprised an initial correction for motion using SLOMOCO2 Beall & Lowe, 2014, detrending of 6th order Legendre polynomials and normalization to signal percentage change (SPC) with AFNI. Furthermore, a mask of white matter voxels was computed from the anatomical image with 3dSeg, which was then eroded 2 voxels to delimit voxels in deep white matter in the functional space. The preprocessed fMRI data were analyzed with three different methods: 1) a traditional general linear model (GLM) analysis using the onsets and durations of the tapping events; 2) the original SPFM approach (3dPFM) using the LASSO for deconvolution and selection of the regularization parameter based on the Bayesian Information Criterion (BIC) Gaudes et al., 2013; and 3) the novel stability-based SPFM with and without the integration operator in its formulation. Both SPFM approaches used the double-gamma canonical HRF as a model for deconvolution (SPMG1 shape in 3dDeconvolve in AFNI). Previous to the final debiasing step, spatio-temporal clustering of a minimum of 5 contiguous voxels with activation (i.e., non-zero coefficient after thresholding) in a temporal window of TR was also performed to remove spurious, scattered activations.
Results and Discussion¶
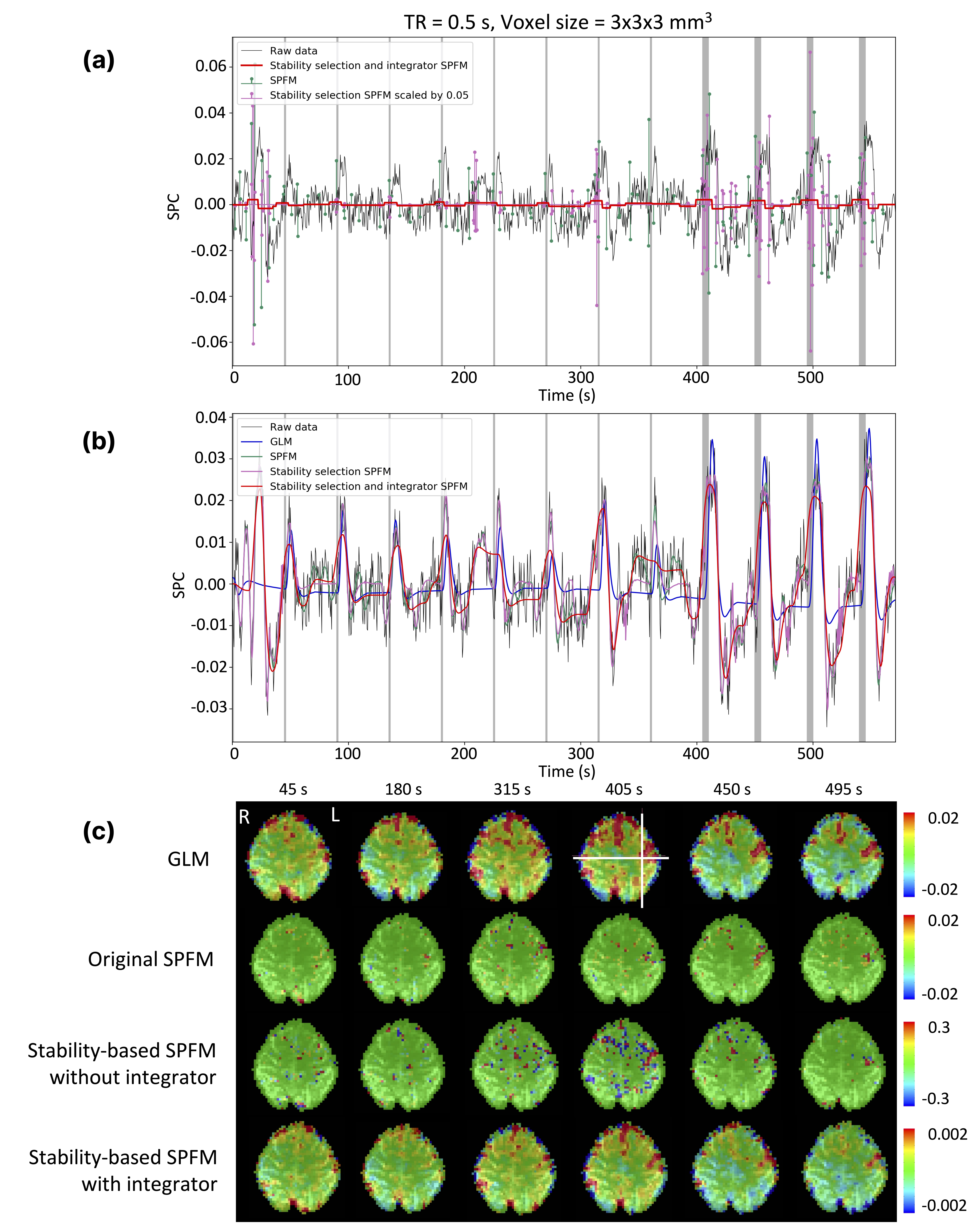
Figure 2:Comparison of the novel stability-based SPFM approach with the SPFM and the GLM methods for data with TR = 0.5 s and a voxel size of iso. (a) plots the time series of the voxels marked with a cross in (c) containing the raw data and the estimates of the different methods as shown in the legend. Onsets and duration of the finger-tapping are depicted as gray vertical lines. (b) shows the estimates of the different methods fitted with the canonical HRF. (c) shows the estimated maps of each of the methods for different finger-tapping events.
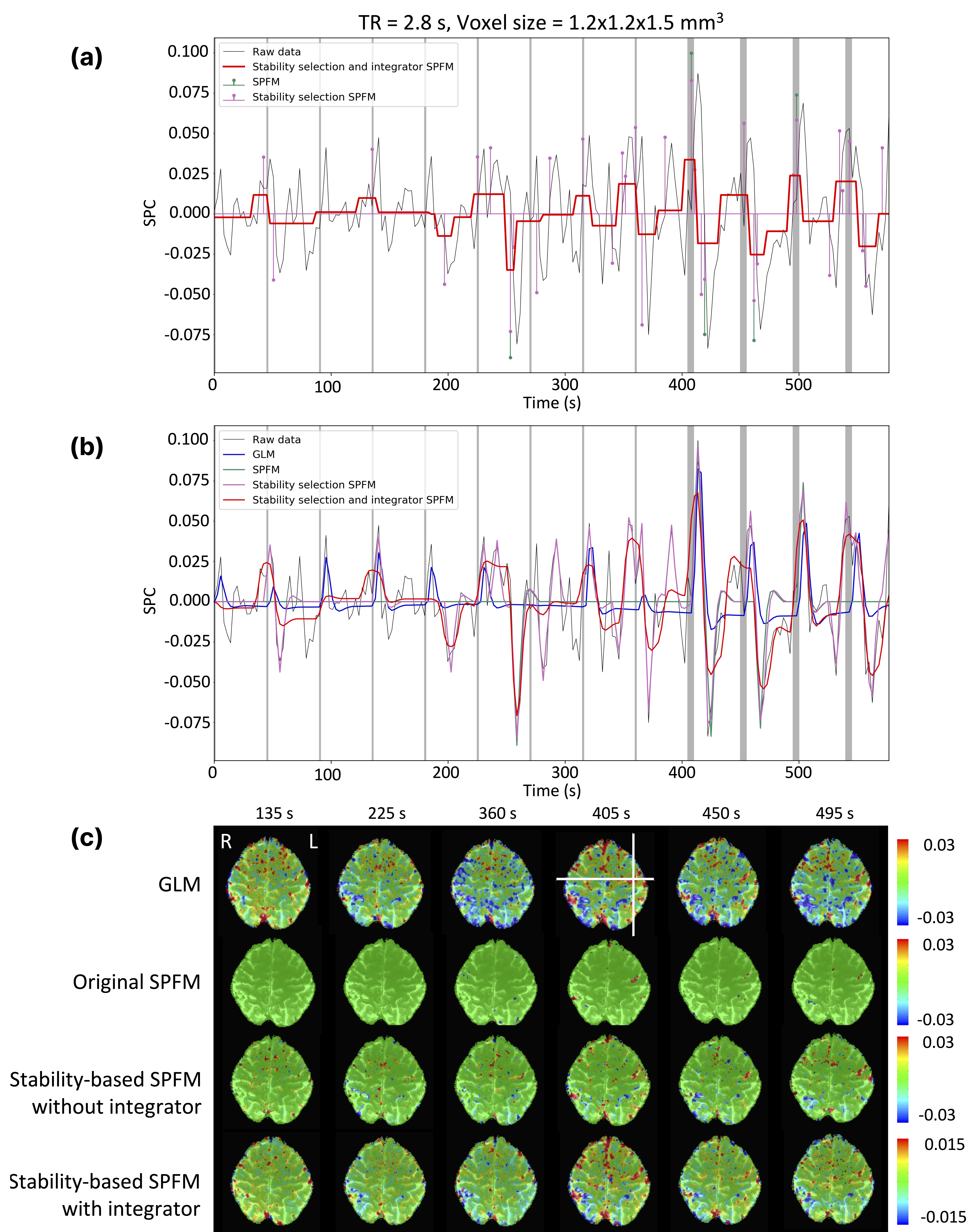
Figure 3:Comparison of the novel stability-based SPFM approach with the SPFM and the GLM methods for data with TR = 2.8 s and a voxel size of . (a) plots the time series of the voxels marked with a cross in (c) containing the raw data and the estimates of the different methods as shown in the legend. Onsets and duration of the finger-tapping are depicted as gray vertical lines. (b) shows the estimates of the different methods fitted with the canonical HRF. (c) shows the estimated maps of each of the methods for different finger-tapping events.
Figure 2 and Figure 3 depict the activity maps estimated with all of the methods for different representative finger-tapping instants and the time courses of a voxel in the left primary motor cortex (marked with a white cross in the maps) for the high temporal and low spatial resolution dataset Figure 2 (a, b and c) and the low temporal and high spatial resolution dataset Figure 3 (a, b and c).
In the high temporal and low spatial resolution scenario (i.e., a high contrast-to-noise ratio regime), the activity maps in Figure 2 (c) illustrate that the original SPFM is able to detect finger tapping events with a high specificity. Implementing stability selection on the original SPFM algorithm increases the sensitivity while maintaining the specificity. However, as it can be seen in Figure 2 (a), the lack of an integration operator yields very variable estimates of the neuronal-related signal after debiasing with least squares (here scaled by 0.05 for visualization purposes) due to the large correlation of the debiasing model with contiguous non-zero coefficients at this fast temporal resolution. Conversely, the novel stability-based SPFM with the integration operator shows activity maps that are comparable to the ground truth despite the lower amplitude of the estimates. Yet, it can be observed that the signal model with the integrator overestimates the duration of the piecewise constant estimates for the short finger tapping events. Thus, in this scenario, the use of the stability selection and the innovation signal exhibits a similar performance to the original SPFM algorithm using LASSO and BIC since the high SNR and high temporal resolution (TR = 0.5 s) enables a precise and clear characterization of the dynamics of the BOLD signal, which facilitates the differentiation between noise and neuronal-related signal.
In an acquisition with a high spatial resolution and a low temporal resolution (i.e., a low contrast-to-noise ratio regime), Figure 3 (a), (b) and (c) demonstrate that the novel stability-based SPFM approach is able to detect more finger-tapping events and their associated brain activity than the original SPFM method. This advantage is clearly seen in the case of the single-tapping events, which exhibit a lower amplitude in the response than the long events with ten consecutive finger taps. The stability selection proves to be essential in correctly estimating finger tapping events, regardless of the use of the integration operator. The addition of the integration operator to the SPFM model produces activity maps that are closer to the ground truth of the GLM analysis (see Figure 2 c), even though the duration of the piecewise constant estimates is overestimated (see Figure 3 a). In this regime, the BIC criterion in the original SPFM is not able to discern between neuronal-related events and noise, failing to detect the finger tapping events, probably as the shape of the BOLD response, which takes 4-6 s to reach its maximum amplitude, cannot be properly characterized by the model owing to the low temporal resolution (TR = 2.8 s). Hence, the stability selection procedure exhibits a robust performance at correctly estimating the neuronal-related events resulting from the finger tapping tasks, which showcases that the additions to the SPFM technique are promising, especially in low temporal resolution settings.
- Uruñuela, E., Jones, S., Crawford, A., Shin, W., Oh, S., Lowe, M., & Caballero-Gaudes, C. (2020, July). Stability-Based Sparse Paradigm Free Mapping Algorithm for Deconvolution of Functional MRI Data. 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). 10.1109/embc44109.2020.9176137
- Gitelman, D. R., Penny, W. D., Ashburner, J., & Friston, K. J. (2003). Modeling regional and psychophysiologic interactions in fMRI: the importance of hemodynamic deconvolution. NeuroImage, 19(1), 200–207. 10.1016/s1053-8119(03)00058-2
- Khalidov, I., Fadili, J., Lazeyras, F., Van De Ville, D., & Unser, M. (2011). Activelets: Wavelets for sparse representation of hemodynamic responses. Signal Processing, 91(12), 2810–2821. 10.1016/j.sigpro.2011.03.008
- Karahanoğlu, F. I., Caballero-Gaudes, C., Lazeyras, F., & Van De Ville, D. (2013). Total activation: fMRI deconvolution through spatio-temporal regularization. NeuroImage, 73, 121–134. 10.1016/j.neuroimage.2013.01.067
- Hernandez-Garcia, L., & Ulfarsson, M. O. (2011). Neuronal event detection in fMRI time series using iterative deconvolution techniques. Magnetic Resonance Imaging, 29(3), 353–364. 10.1016/j.mri.2010.10.012